Authors: Elspeth A. Bruford, Bryony Braschi, Paul Denny, Tamsin E. M. Jones, Ruth L. Seal, Susan Tweedie
Synopsis
This paper describes how genes (protein-coding, pseudogenes, and non-coding RNAs) are named. Pretty straightforward.
Summary
Naming protein-coding genes
A gene is a DNA segment that contributes to phenotype/function. Most protein-coding genes are named after their function. Genes with a common root symbol typically form some gene group that have sequence homology, shared function, or shared membership in a protein complex. Gene suffixes can be Arabic numerals, letters, or some combination of the two.
Genes with unknown function are named in 5 ways:
- Recognized structural domain and motifs encoded by the gene (E.g. ABHD1, “abhydrolase domain containing 1”; HEATR1, “HEAT repeat containing 1”)
- Homolog with other gene in human genome with known function (e.g. CASTOR3 is “CASTOR family member 3”). The placeholder FAM (“family with sequence similarity”) is used when the function of none of the homologous genes have known function. Each family has a unique FAM number (FAM3) and each family member has unique letter or letter and number (FAM3A or FAM4C2P).
- Homolog with gene in another species. If the gene has a one-to-one ortholog, it can take the exact name in other species (CDC45 for “cell division cycle 45” in yeast). A unique number or letter suffix added if there is more than one human homolog.
- Presence of open reading frame. Genes that don’t fit the previous 3 criteria are designated: chromosome, “orf” in lowercase, and the number in a series (eg. C3orf18, “chromosome 3 open reading frame 18”). If the gene’s coding potential is uncertain, the word “putative” is added in the name (“chromosome 18 putative open reading frame 15”).
- Genes identified by Human cDNA project at Kazusa DNA Research Institute are named using KIAA# identifiers
Naming pseudogenes
A pseudogene is a section of a chromosome that is incapable of producing a functional protein product, but has a high level of homology to a functional gene. Usually they are named after their parent with a “P” suffix (e.g. DPP3P1, “DPP3 pseudogene 1”; CBWD4P, “COBW domain containing 4, pseudogene”). The parent gene can be a functional ortholog in another species (e.g. ADAM24P, “ADAM metallopeptidase domain 24, pseudogene” after mouse Adam24). Sometimes pseudogenes won’t include the “P” if the symbol is well established (e.g. UOX, “urate oxidase (pseudogene)”). Some genes are pseudogenes in the reference and coding in some of the population. These genes are classified as protein-coding but indicated by “(gene/pseudogene)”
Naming non-coding RNAs
Non-coding RNAs (ncRNAs) are named according to their type (miRNA, tRNA, etc.). Long non-coding RNAs (lncRNAs) are named according to a key function or characteristic of the RNA, otherwise the following flowchart:
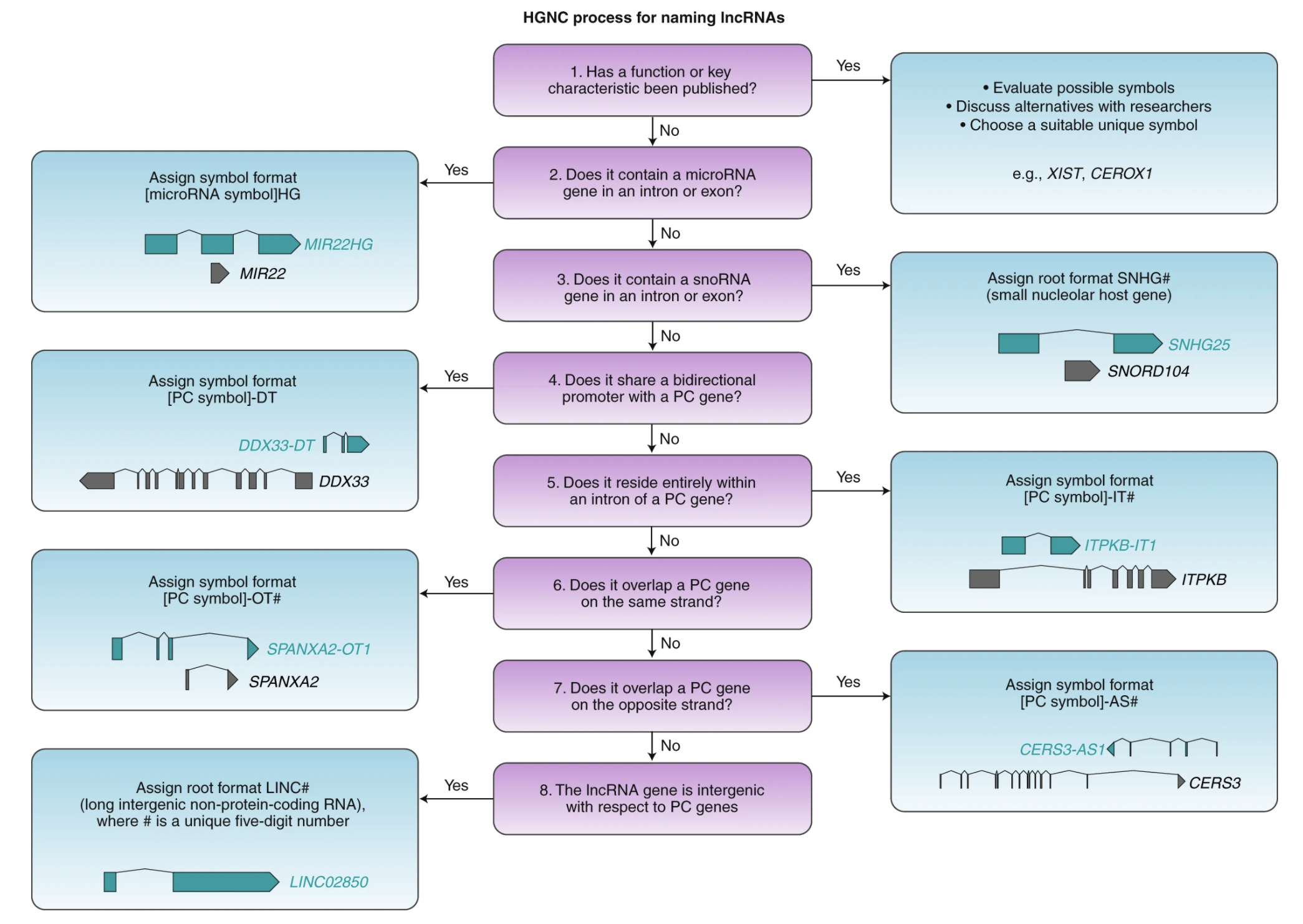
HG, host gene; PC, protein coding; DT, divergent transcript (used for lncRNA genes that share a promoter with a PC gene); IT, intronic transcript; OT, overlapping transcript; AS, antisense RNA; LINC, long intergenic non-protein-coding RNA
Naming readthrough transcripts
Readthrough transcripts are named by two symbols separated by a hyphen (e.g. INS-IGF2).